Quick ggplot2 Tip: Creating Duplicate Legends
Duplicate a plot legend's to add a second key where the audience can better understand your figure
Mastering the R package ggplot2
The ggplot2 package provides powerful methods to display data as
graphics. The beauty of the package lies in it’s simplicity -
understanding the core methods (applying variables to aesthetics and
transformations) covers ~95% of static visualizations a data
visualization developer might be interested in generating. Most of final
5% can be achieved by understanding the infrastructure of the package.
One such example is how plot components are “written” to the graphics
device.
I’ll walk through how legends are generated and how we can create a second duplicate legend to bookend the top and bottom of a long bar plot. I I contributed a method to solve this problem on stackoverflow and wanted to get into some further details in this post.
Let’s walk through creating a second legend in ggplot2.
Creating Dual Legends in ggplot2 - Libraries and Data Reshaping
The midwest data set from the ggplot2 package contains demographic
information of midwest counties and should work as a representative
dummy data set for this post. Here is a quick overview of the midwest
data set using glimpse:
R Libraries, `Midwest` data set overview
# load libraries and fonts
library(ggplot2)
library(scales)
library(dplyr)
library(grid)
library(extrafont)
loadfonts(quiet = TRUE)
glimpse(midwest)I’ll subset the data a little bit to accomodate the plot and organize
the display order of counties by their total population. The
theme_white block of code adds some basic aesthetics and fonts.
R Libraries, Data Manipulation, and Plot Generation
# load libraries and fonts
library(ggplot2)
library(scales)
library(grid)
library(extrafont)
loadfonts()
# data subset and refactoring
midwest <- midwest[!duplicated(midwest$county), ]
midwestAAU <- midwest %>% filter(category == "AAU") %>% arrange(poptotal)
midwestAAU$county <- factor(midwestAAU$county,levels = midwestAAU$county,labels = toupper(midwestAAU$county))
gg <- ggplot(midwestAAU) + geom_bar(aes(y = poptotal, x = county, fill = state), stat = "identity") +
scale_y_continuous(expand = expand_scale(mult = c(0, .1)), labels = comma) + coord_flip() + theme_minimal() + labs(title = "Random Midwest Counties Arranged by Total Population",y = "Total Population", x = "County", fill = "State") + scale_fill_brewer(palette = "Set3")
# plot aesthetics
theme_white <- theme(text = element_text(family="Open Sans"),
panel.grid.major.y=element_blank(),
panel.grid.major.x=element_blank(),
panel.grid.minor.x=element_blank(),
panel.grid.minor.y=element_blank(),
plot.title=element_text(size=24,family = "Open Sans",lineheight=.75),
axis.title.x=element_text(size=20, family = "Open Sans Semibold"),
axis.title.y=element_text(size=20,family = "Open Sans Semibold"),
axis.text.x=element_text(size=12),
axis.text.y=element_text(size=12),
axis.ticks = element_blank(),
legend.position = "bottom",
legend.margin = margin(b = 0)
)
# apply theme and export plot
gg <- gg + theme_white
ggsave(gg, filename = "midwestPlot.png",height = 12, width = 12, dpi = 300, units = "in", device='png')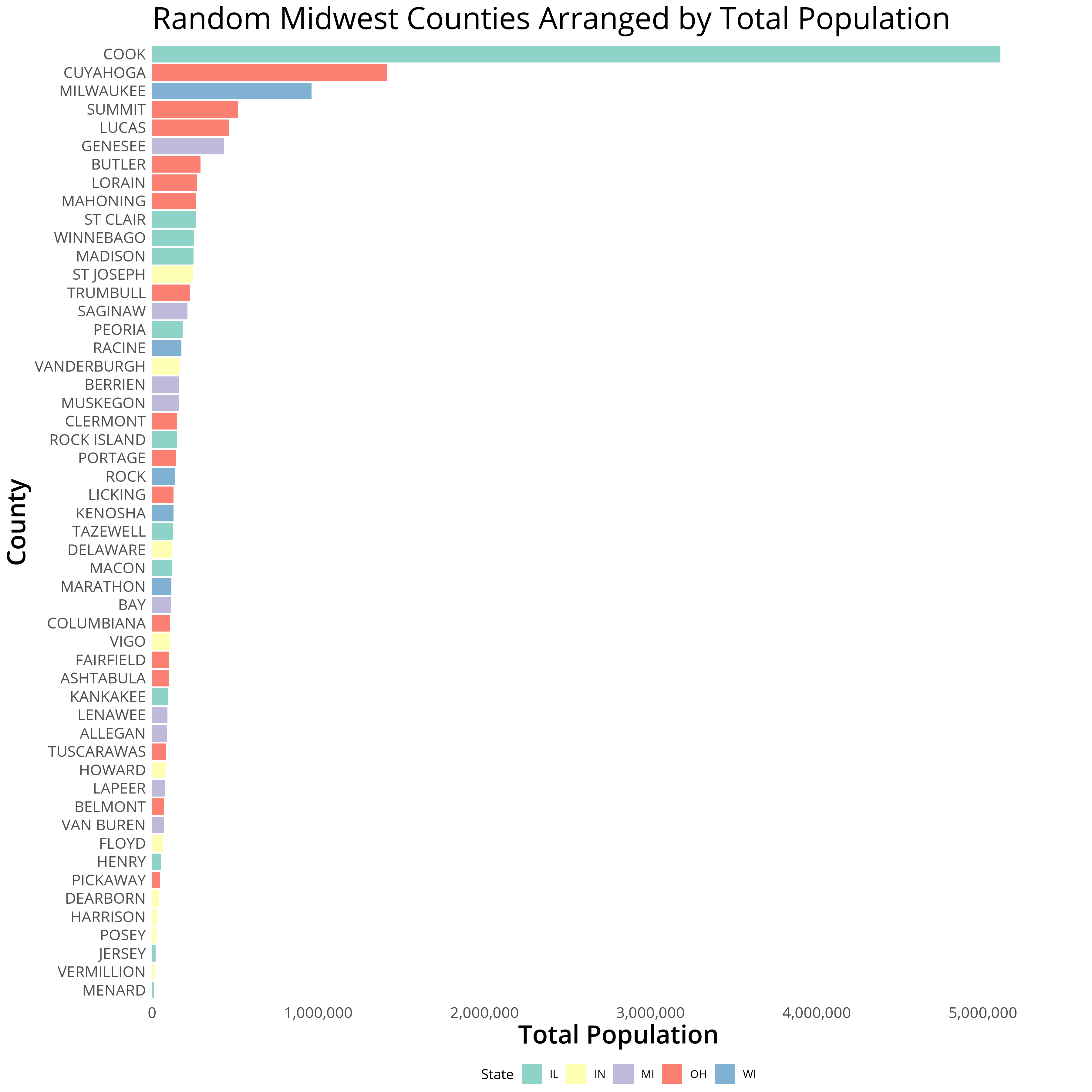
Now that we’ve generated our plot we can focus on creating the second legend.
Creating a Second Legend in ggplot2
So far we’ve covered ggplot2 functionalities that should create the
~95% of plots I discussed earlier. To expand upon these, let’s get into
some ggplot2 internals. The function ggplotGrob allows us to parse
our saved gg graphical object. This object can be manipulated to
override default ggplot2 conventions or provide methods to hack our
plot in ways that the package isn’t designed for intentionally
(i.e. where there isn’t a compiled function.)
The createTopLegend function below easily duplicates a bottom legend
at the top of the plot by:
- Grabbing the ggplot graphical object
- Retrieving the legendGrob within the ggplot object
- Duplicating the legendGrob layout
- Specifying the location of the new legendGrob
- Appending the new legendGrob to the ggplot object
createTopLegend Function
createTopLegend <- function(ggplot, heightFromTop = 1) {
# grab the saved ggplot2 object
g <- ggplotGrob(ggplot)
# count the number of grobs in this plot (which we'll use to append another)
nGrobs <- (length(g$grobs))
# find the guide-box object which provides the plot information for the legend
legendGrob <- which(g$layout$name == "guide-box")
# duplicate the legend's grob and layout
g$grobs[[nGrobs+ 1]] <- g$grobs[[legendGrob]]
g$layout[nGrobs+ 1,] <- g$layout[legendGrob,]
# g$layout$t <- ifelse( g$layout$t > heightFromTop, g$layout$t + 1, g$layout$t)
# retrieve the alignment of the legend
rightLeft <- unname(unlist(g$layout[legendGrob, c(2,4)]))
# specify the location of the new legendGrob (t,r,b,l)
# use the heightFromTop argument to adjust the vertical positioning
g$layout[nGrobs+ 1, 1:4] <- c(heightFromTop, rightLeft[1], heightFromTop, rightLeft[2])
g
}We can then apply the createTopLegend function on our saved ggplot2
object gg and redraw our plot with grid.draw:
Plot with Duplicate Legend…Overlapping
gg2 <- createTopLegend(gg, 3)
ggsave(grid.draw(gg2), filename = "midwestPlot2.png",height = 12, width = 12, dpi = 300, units = "in", device='png')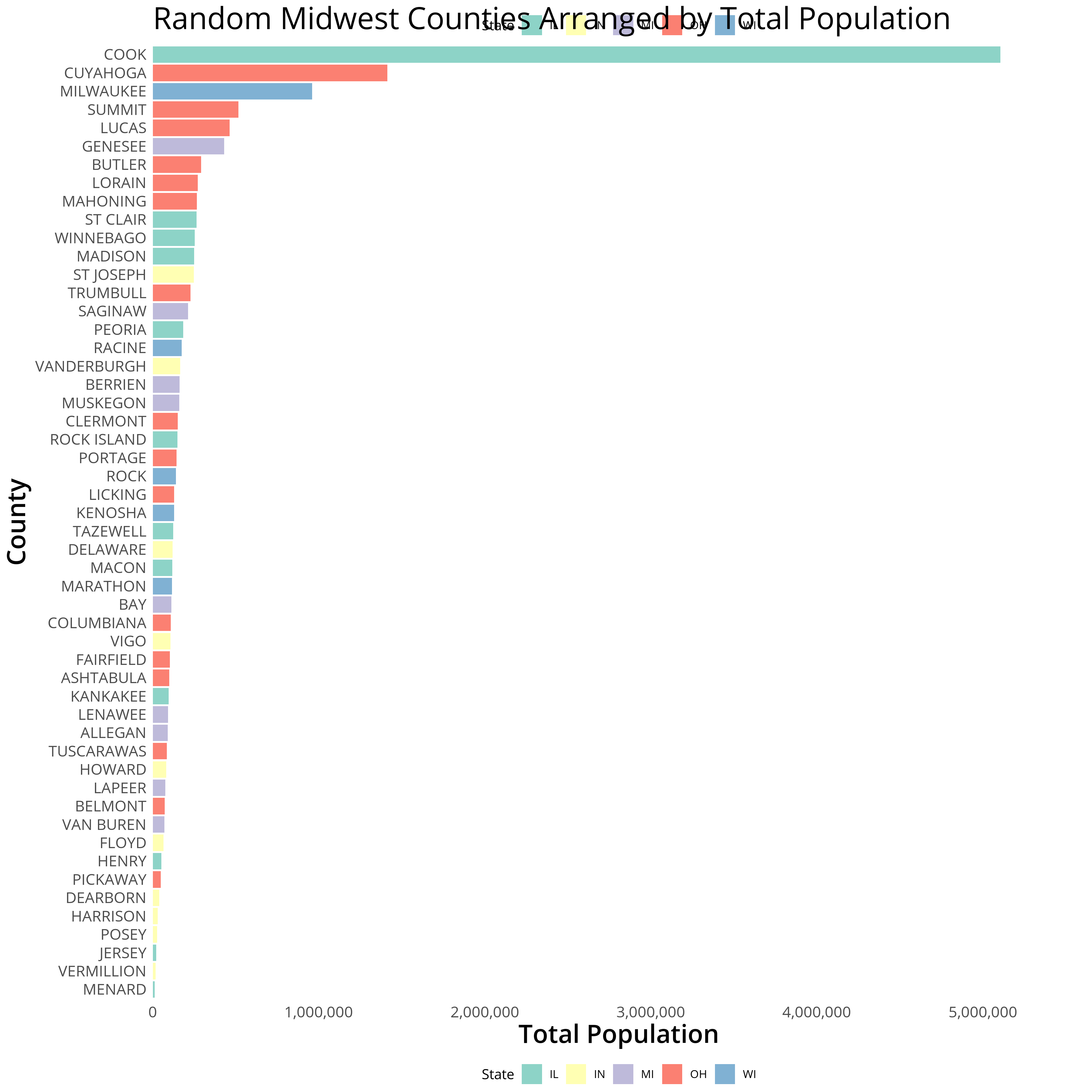
You’ll notice that our top legend now overlaps with the positioning of
the title. To remedy this we can add some additional margins from within
theme_white. We’ll add a bottom margin to the title to add spacing, a
bottom margin to the legends, and a negative margin to the bottom of the
plot. Each of these margins work in tandem so the negative plot margin
is necessary to account for the extra spacing we’re adding to the top
legend for the plot to be appropriately spaced.
Fiddling with Title, Legend, and Plot Margins to Accommodate for the Top Legend
# plot aesthetics
theme_white <- theme_white + theme(
plot.title=element_text(size=24,family = "Open Sans",lineheight=.75, margin = margin(b = 40)),
legend.margin = margin(b = 40),
plot.margin = margin(t = 10, r = 10, b = -30, l = 10)
)
gg <- gg + theme_white
gg2 <- createTopLegend(gg, heightFromTop = 4)
ggsave(grid.draw(gg2), filename = "midwestPlotLegend.png",height = 12, width = 12, dpi = 300, units = "in", device='png')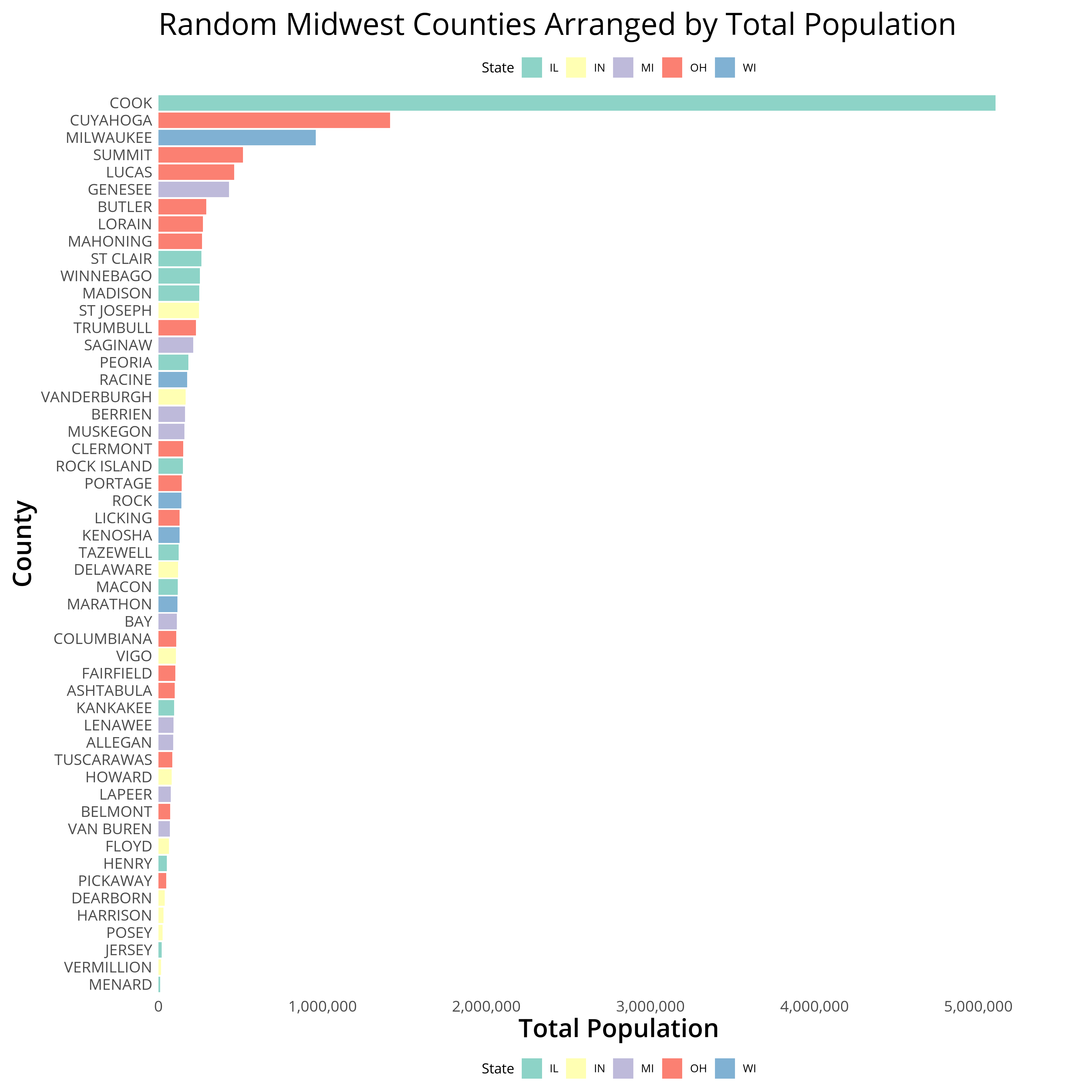
Interested in learning more? Hire me to consult on your next project, follow me on twitter, or contact me via email. All inquiries welcome!